Metadata
The “Metadata” module serves as your “dashboard” and “toolbox” when you run analyses on SeekSoulOnline. It consolidates key capabilities to keep every workflow clear, traceable, and efficient. The module is composed of two core sections:
- Label Management: Offers a full suite of metadata (meta.data) editing tools, enabling you to flexibly create, modify, merge, and manage cell labels to uncover deeper insights.
- Operation Log: Faithfully records every action you take on meta.data within the platform, providing a complete history for troubleshooting and auditing.
NOTE
The Metadata module helps trace analysis workflows, label changes, and operation histories. Review it regularly to maintain data consistency.
Label Management
This section introduces tools that help you manage and manipulate metadata (meta.data) labels for cells. You can delete, edit, merge, and add labels to satisfy diverse analysis needs.
Within the current workflow you can delete, edit, merge, and add labels. Click “x” to delete a column, and click “✏” to edit it.
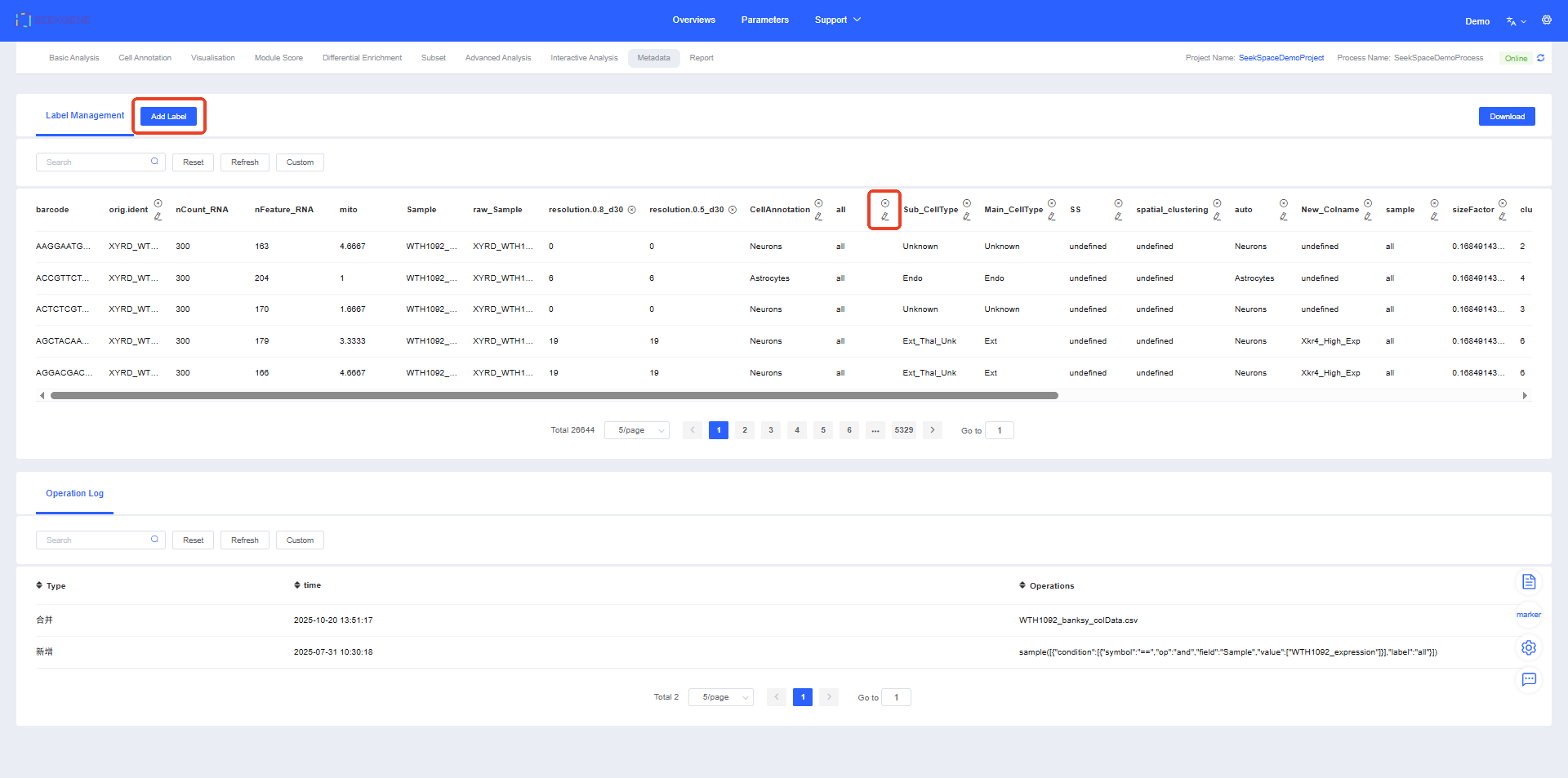
Click Add Label to merge or create labels.
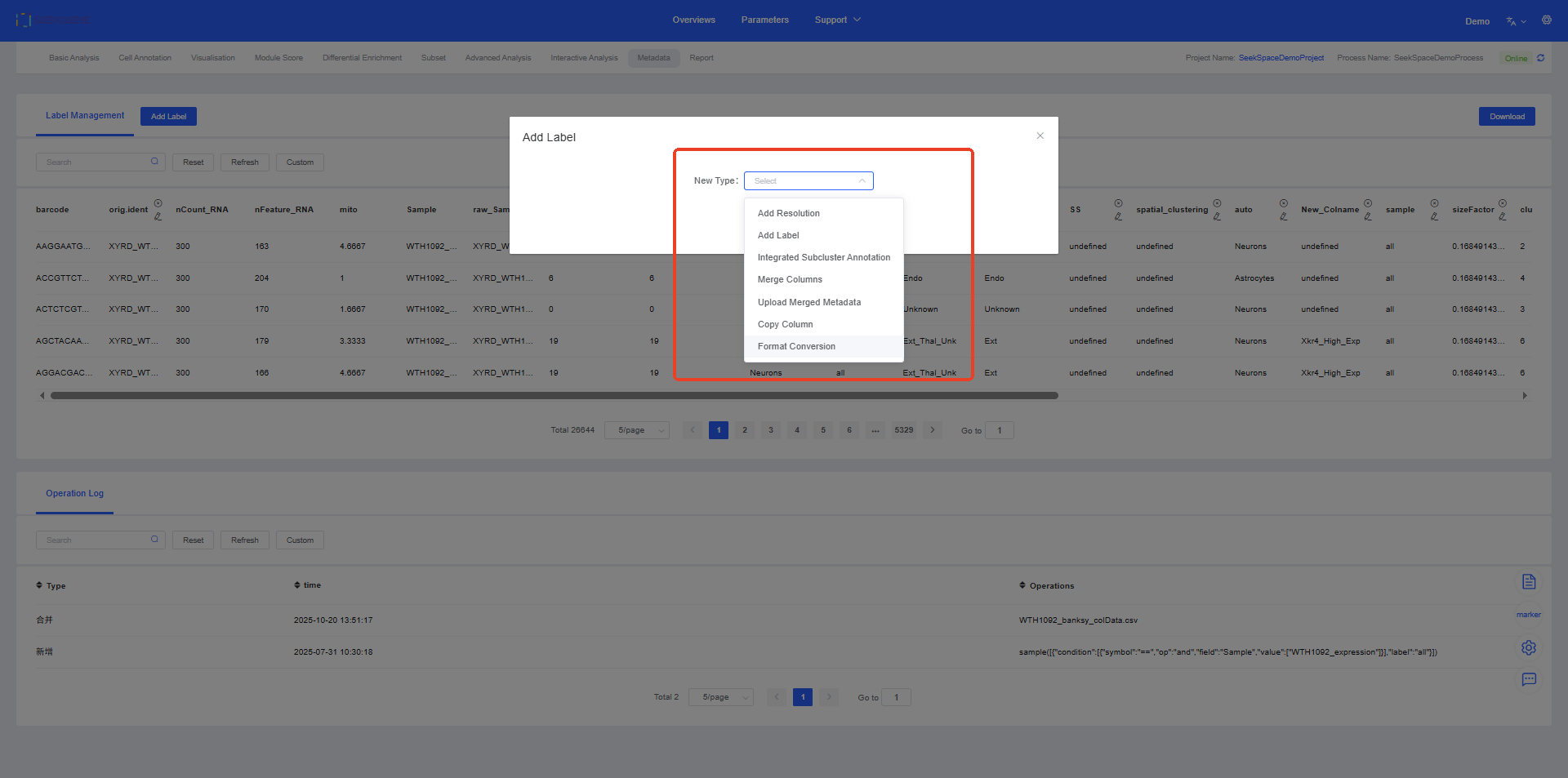
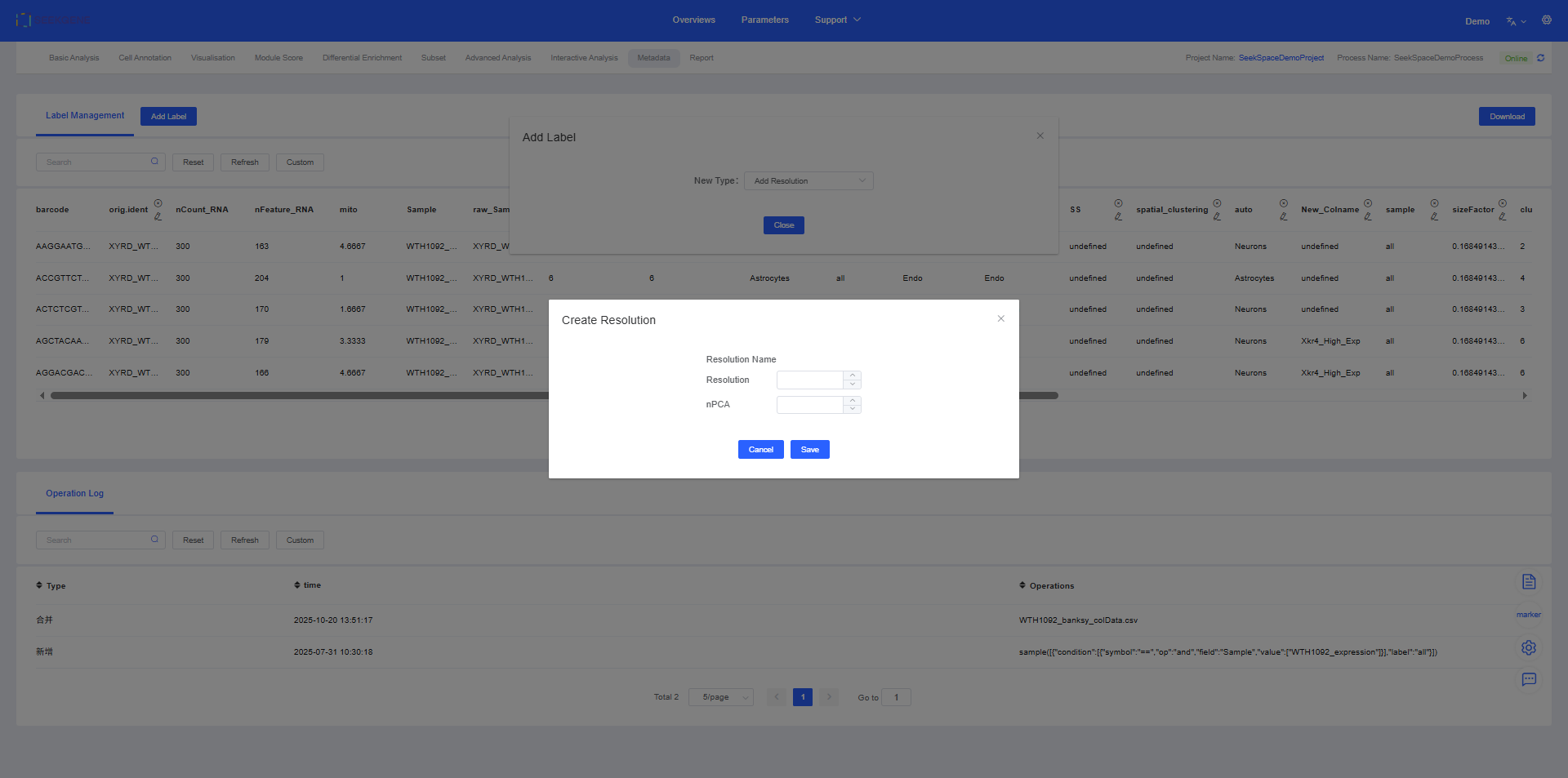
1. Add Resolution
The “Add Resolution” feature allows you to adjust the granularity of clustering, affecting the fineness of cell grouping. This is consistent with the functionality in the Cell Annotation module.
Core Advantages:
- Explore cell structures at different levels: By adjusting the resolution, you can explore cell populations at different scales. Higher resolutions produce more, smaller cell clusters, suitable for fine-grained analysis; lower resolutions produce fewer, larger cell clusters, suitable for macroscopic observation.
- Optimize analysis strategies: Choose the most suitable resolution based on your biological questions to obtain the best cell grouping results.
This feature provides great flexibility for exploratory data analysis, helping you discover biological patterns at different levels.
2. Add Label
The “Add Label” feature allows you to group and tag cells (Barcode) based on one or more conditions, creating new custom labels. This is useful for filtering, identifying, and analyzing specific cell subpopulations from complex data. The functionality is consistent with the “Add Label” module in Visualisation.
Function Advantages
- Precise filtering of cell populations: By setting multiple filtering conditions (e.g., from specific samples, expressing specific genes, belonging to certain clusters), you can accurately define and separate the cell populations of interest.
- Simplify complex analysis: By grouping cells that meet multiple conditions into a new label, you can simplify subsequent differential expression analysis, visualization, and functional enrichment analysis.
- Increase analysis flexibility: You can dynamically create arbitrarily complex cell groupings based on research needs, without being limited to predefined annotations.
Operation Steps
Click Add Label and select the “Add Label” feature.
After entering the “Add Label” page, follow these steps:
- Name the new label: Enter a descriptive name for your new label in the “Name” field.
- Set filtering conditions:
- Choose the Category to filter (e.g.,
seurat_clusters). - Use Operator (“==” equals or “≠” not equals) and Value to define conditions.
- If multiple conditions are needed, use Condition (“and” intersection or “or” union) to combine them.
- Choose the Category to filter (e.g.,
- Define default values: Set a default label for cells that do not meet the conditions, usually “Undefined”, but can be customized.
- Complete addition: Click the Add Label button and the system will create a new label column based on your conditions. This new label can be used in all subsequent analyses and visualizations.
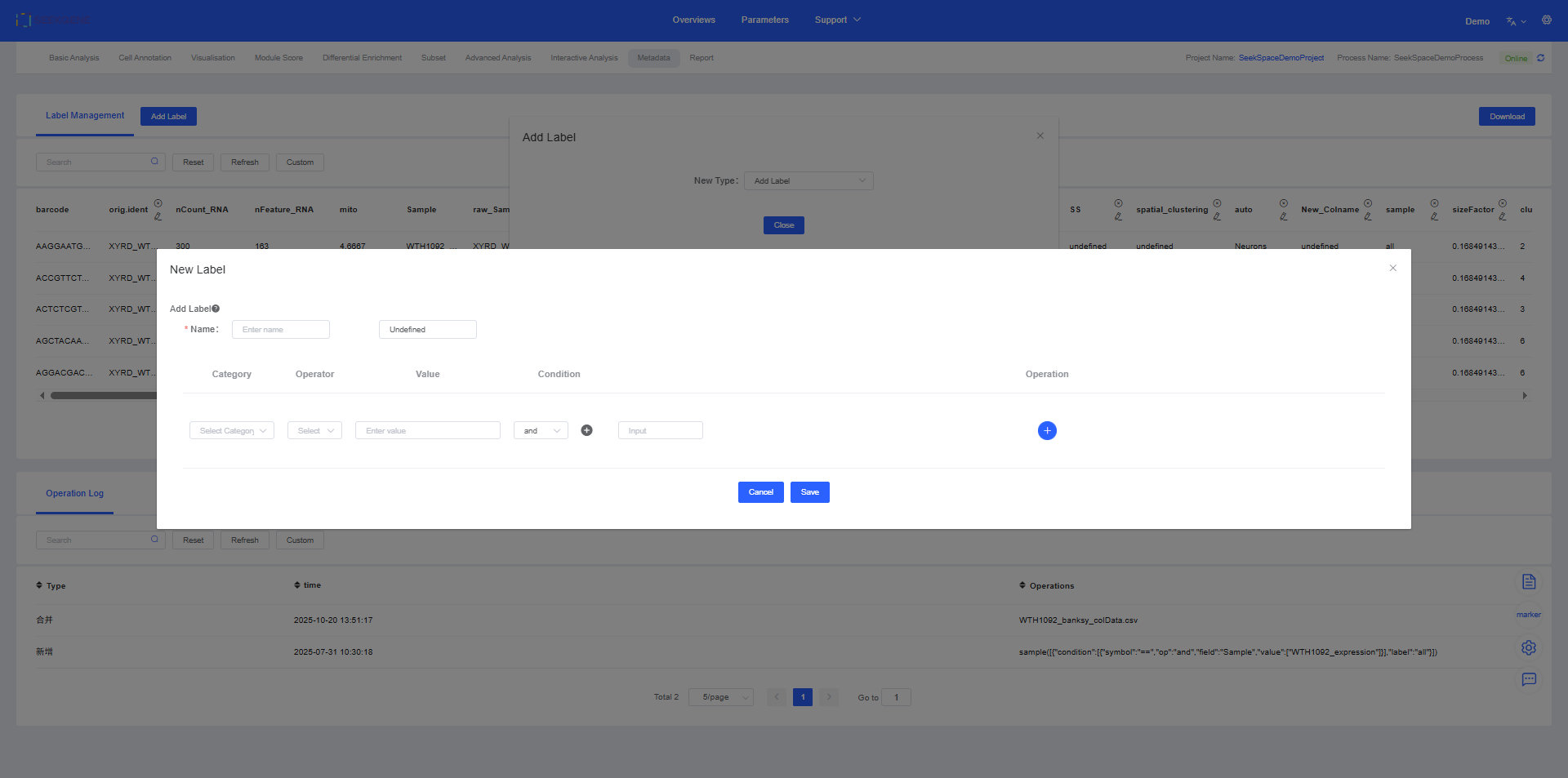
For example, you can create a label to mark all cells from “Sample A” and belonging to “T cells” for specialized analysis.
3. Integrated Subcluster Annotation
In single-cell analysis, we often re-cluster a large cell population (e.g., T cells) to identify finer subgroups (e.g., CD4+ T cells, CD8+ T cells). The “Merge Subgroup Annotations” feature can integrate these finer subgroup labels back into the original cell annotation column, updating and unifying the annotation information.
Application Scenarios:
- Update annotations: After completing fine-grained annotations for a cell subgroup (e.g., B cells), you want to update these detailed labels (e.g., “Memory B cell”, “Plasma B cell”) into the main workflow’s overall cell type labels, replacing the original “B cell” label.
Operation Flow:
- First, select the workflow and label column containing the main cell population (large group).
- Then, select the workflow and label column containing the fine-grained subgroup annotations.
- The system will precisely overlay the subgroup annotations onto the corresponding cells in the large group, generating a new, more informative cell annotation column.
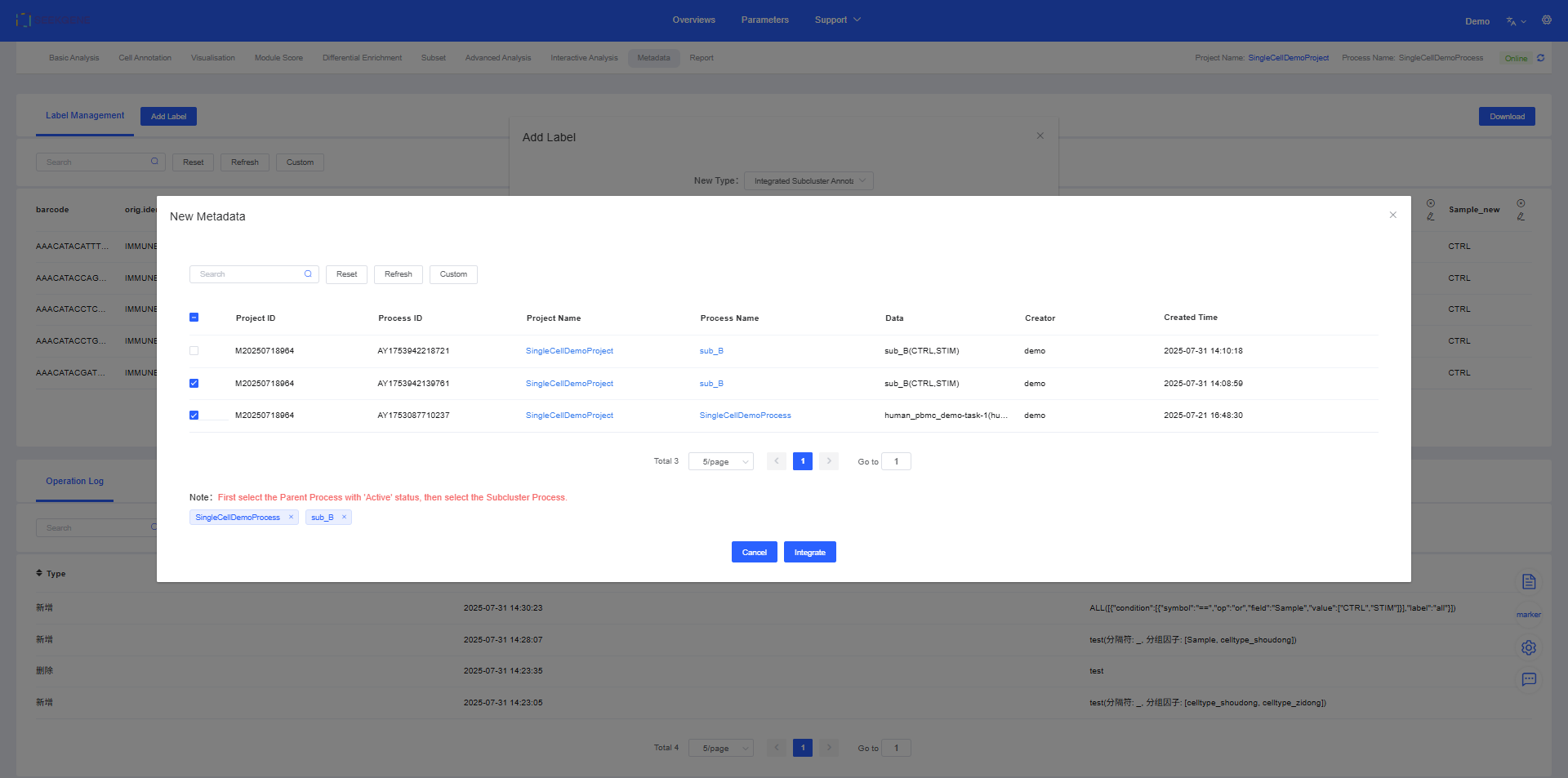
Select the label to be overlaid in the large group (e.g., celltype) and the label to be overlaid in the subgroup (e.g., subtype).
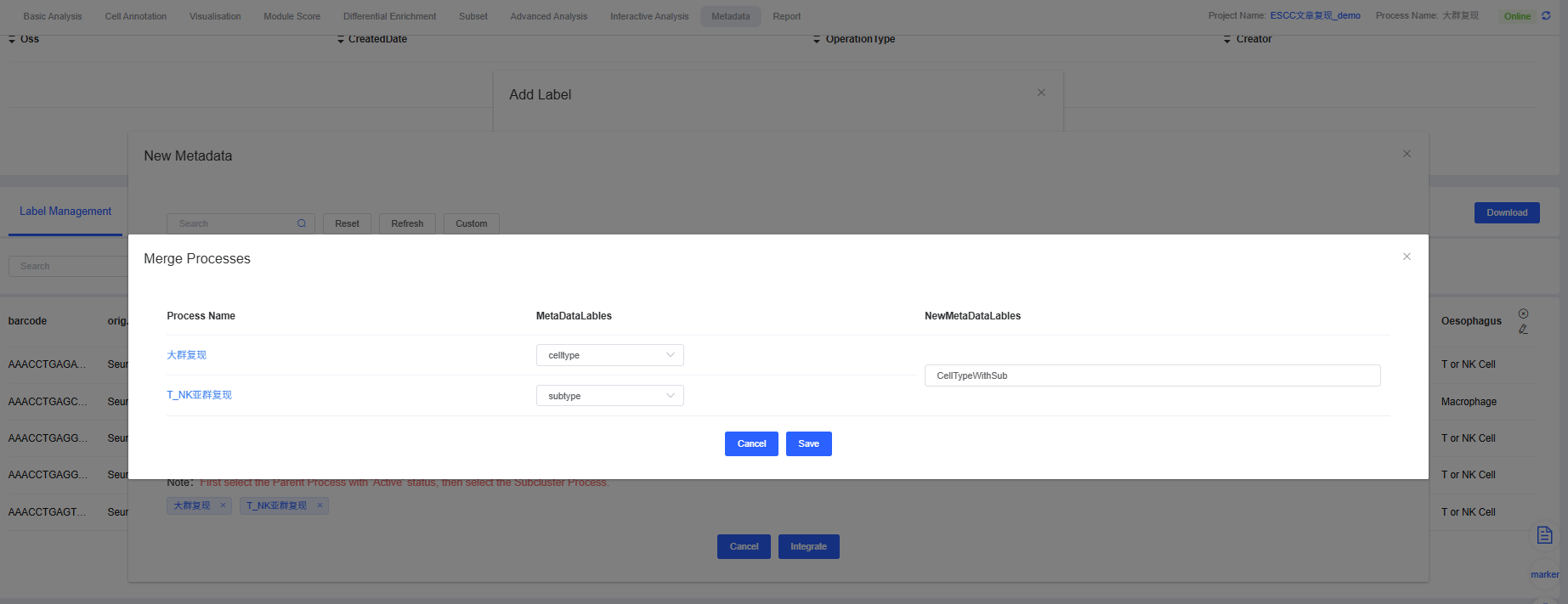
You can see that the label “celltype” in the large group is overlaid with the label “CellTypeWithSub” in the subgroup.
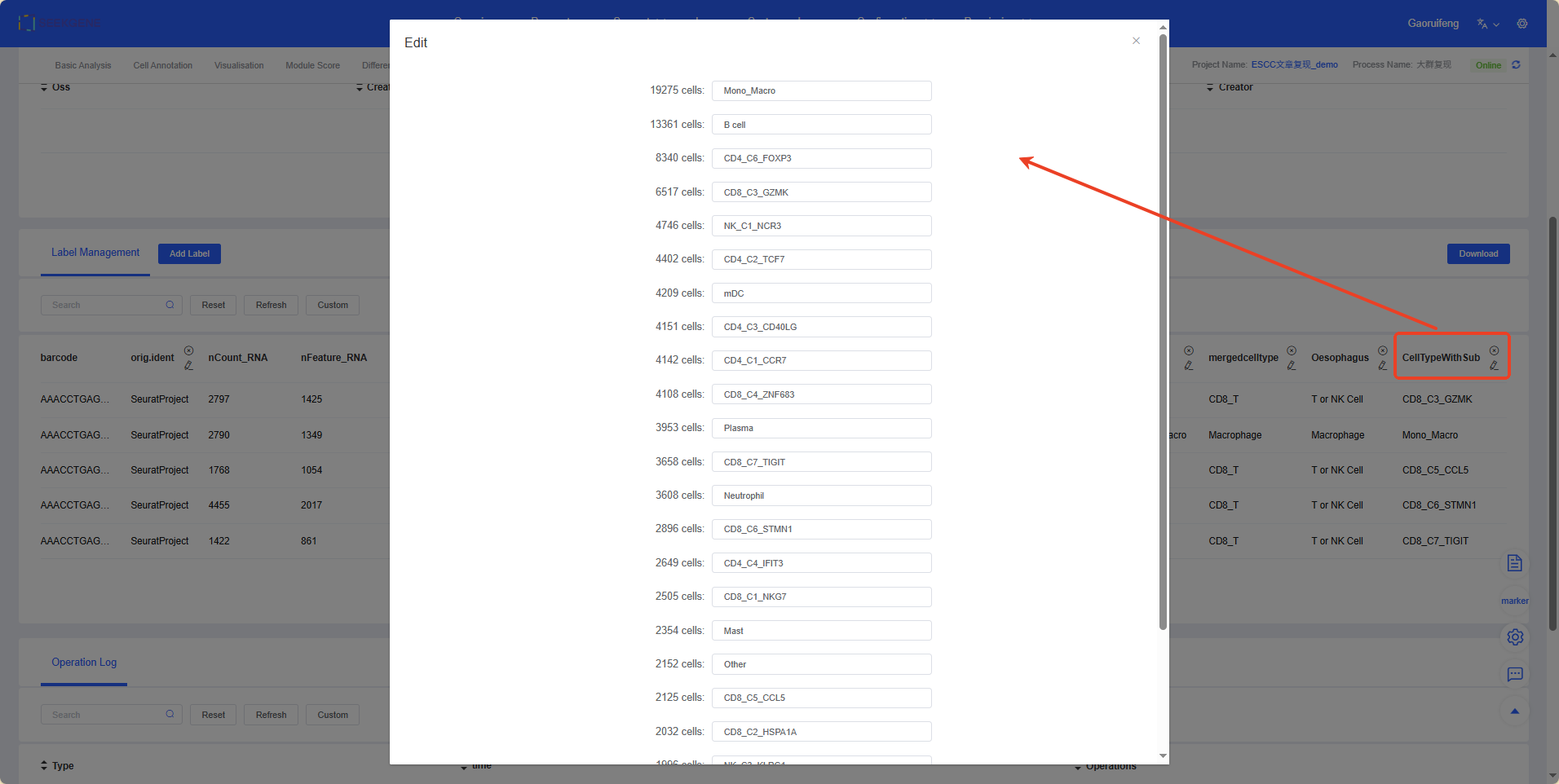
CAUTION
When merging subgroup annotations, the original label content will be overwritten by the new label. Please operate with caution.
4. Merge Columns
The “Merge Columns” feature allows you to combine multiple existing metadata columns (labels) into a single, more informative column. This is useful for creating composite labels that reflect multiple experimental conditions or cell characteristics.
Usage Scenarios:
- Integrate experimental information: Merge “sample source” (e.g.,
Sample) and “treatment condition” (e.g.,Treatment) to generate a unique label for each cell group (e.g.,SampleA_DrugX). - Create fine-grained annotations: Merge a coarse cell type annotation (e.g.,
T_cell) with a finer clustering result (e.g.,cluster_id) to obtain a more accurate cell subgroup definition (e.g.,T_cell_cluster1).
Operation Steps:
- New Tag Name: Specify a descriptive name for the merged column.
- Separator: Select a separator (e.g., “_” or “-”) to connect the contents of the merged columns.
- Group.by: Check the columns you want to merge.
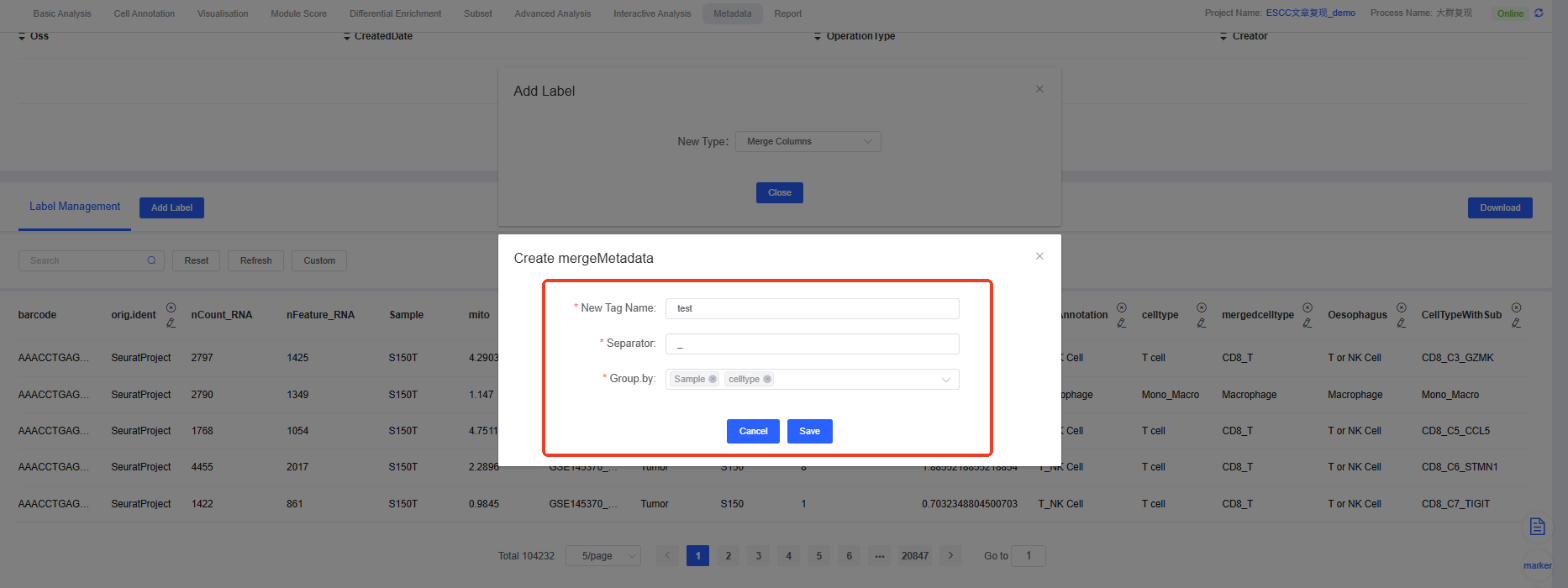
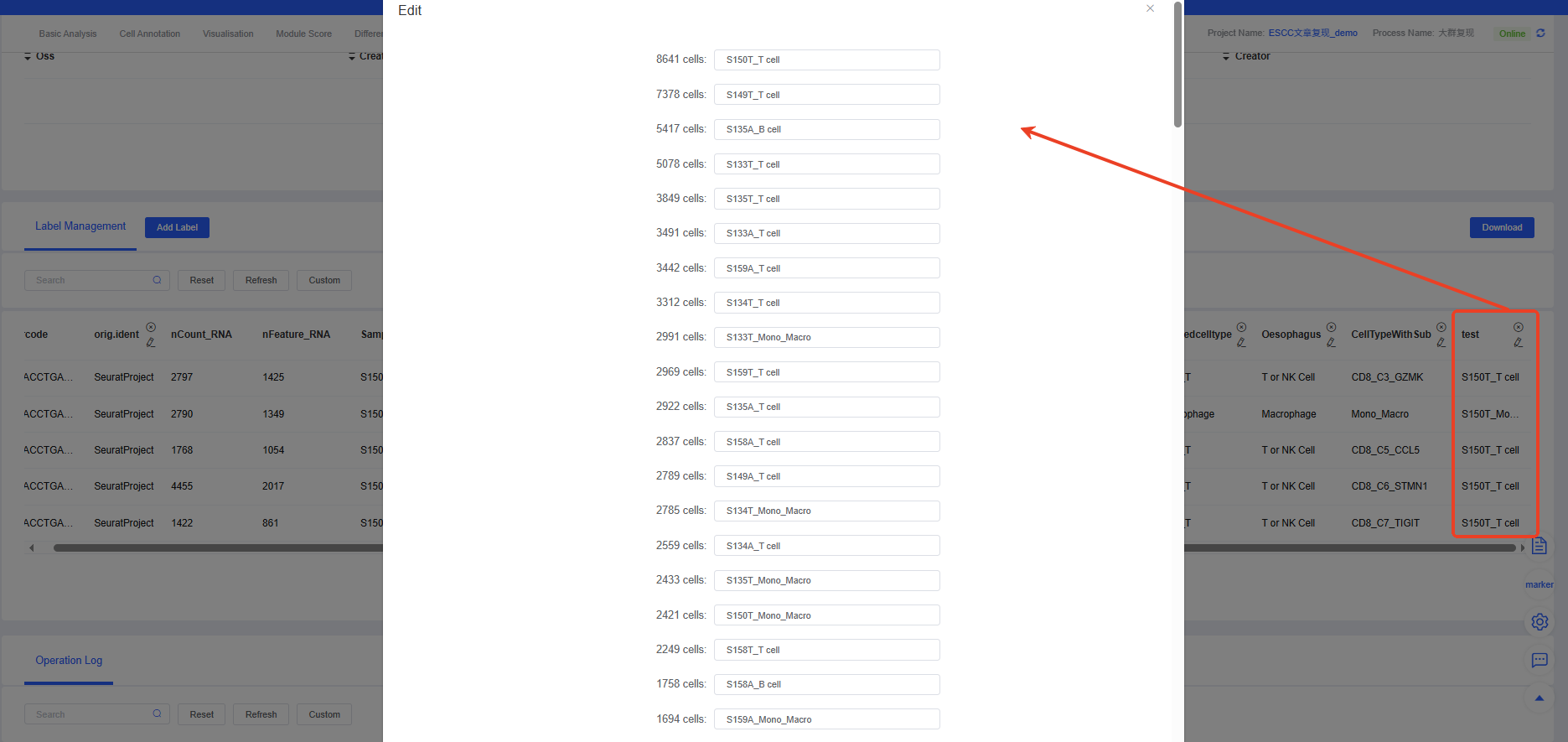
After merging, the new composite label will appear in your metadata, as shown below, where the labels “Sample” and “celltype” are merged into a new label “test” using the separator “_”.
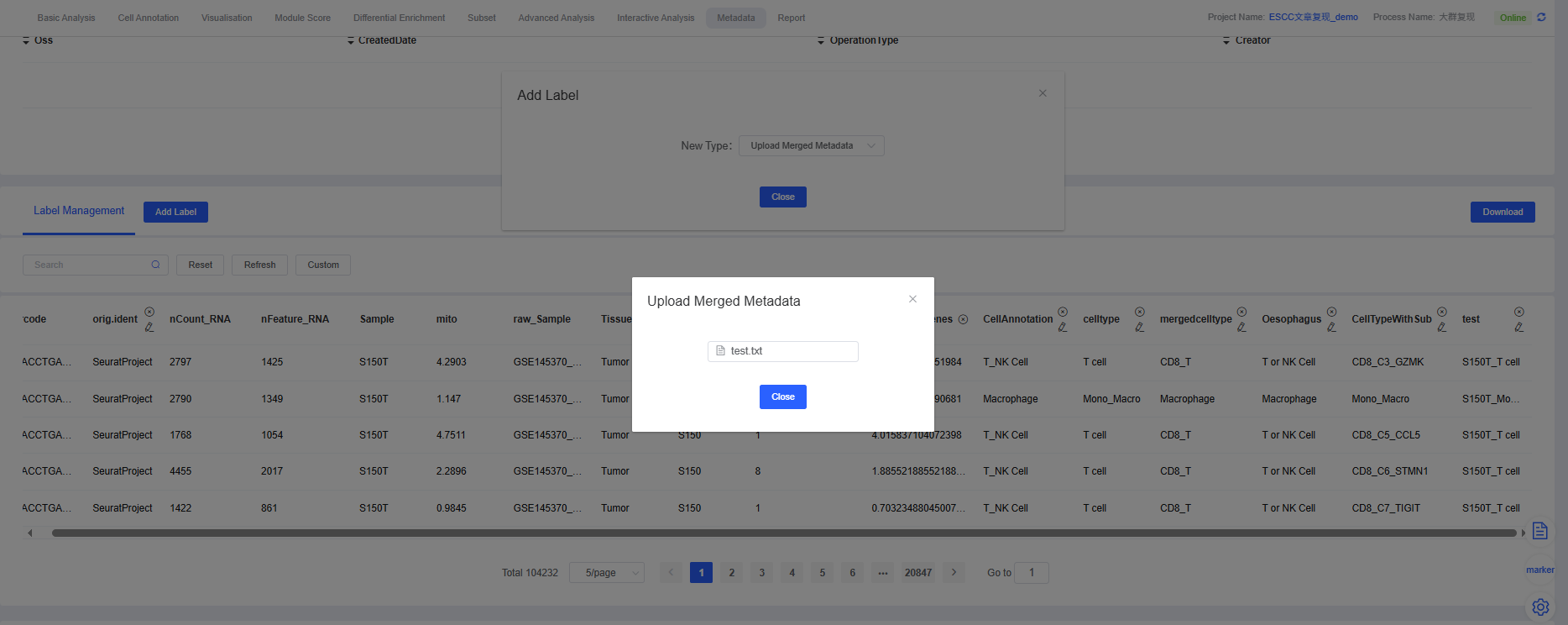
5. Upload Merged Metadata
This feature is crucial for data integration. It allows you to upload and merge local metadata files (e.g., clinical information, sample batches, or annotations generated by other analyses) into the current cloud platform analysis workflow.
Core Requirements:
- File format: Supports
.txtor.csvformats. - Matching key: Your local file must contain a column named
barcode, whose content needs to match the cell barcodes in the cloud platform workflow. This is the basis for merging. - Column name uniqueness: Besides the
barcodecolumn, the local file needs to contain at least one more information column, and other column names cannot conflict with existing metadata column names in the workflow to avoid conflicts.
Application Value:
- Integrate multi-source data: Associate external clinical data, sample information, or custom annotations with single-cell expression data for more in-depth integrated analysis.
- Import analysis results from other tools: Import clustering or annotation results generated by other software (e.g., Seurat, Scanpy) into the platform for cross-platform analysis.
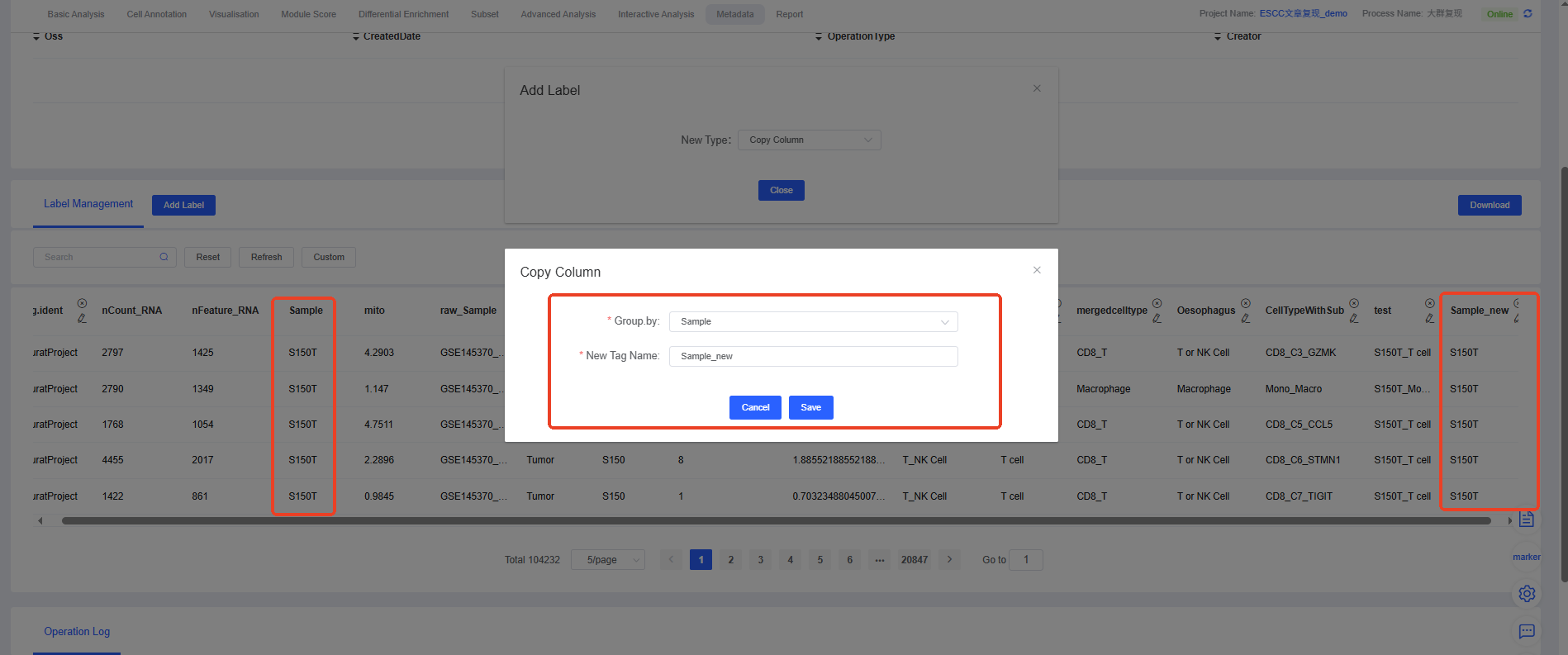
6. Copy Column
The “Copy Column” feature is a simple yet practical tool that allows you to create an exact copy of an existing column and assign it a new name.
Common Use Cases:
- Backup and exploration: Before modifying or reclassifying an important annotation column (e.g., manually annotated cell types), create a backup copy to prevent accidental changes.
- Create derivative labels: Use an existing column as the basis for a new label, then modify the copy (e.g., merge or delete certain categories) without altering the original data.
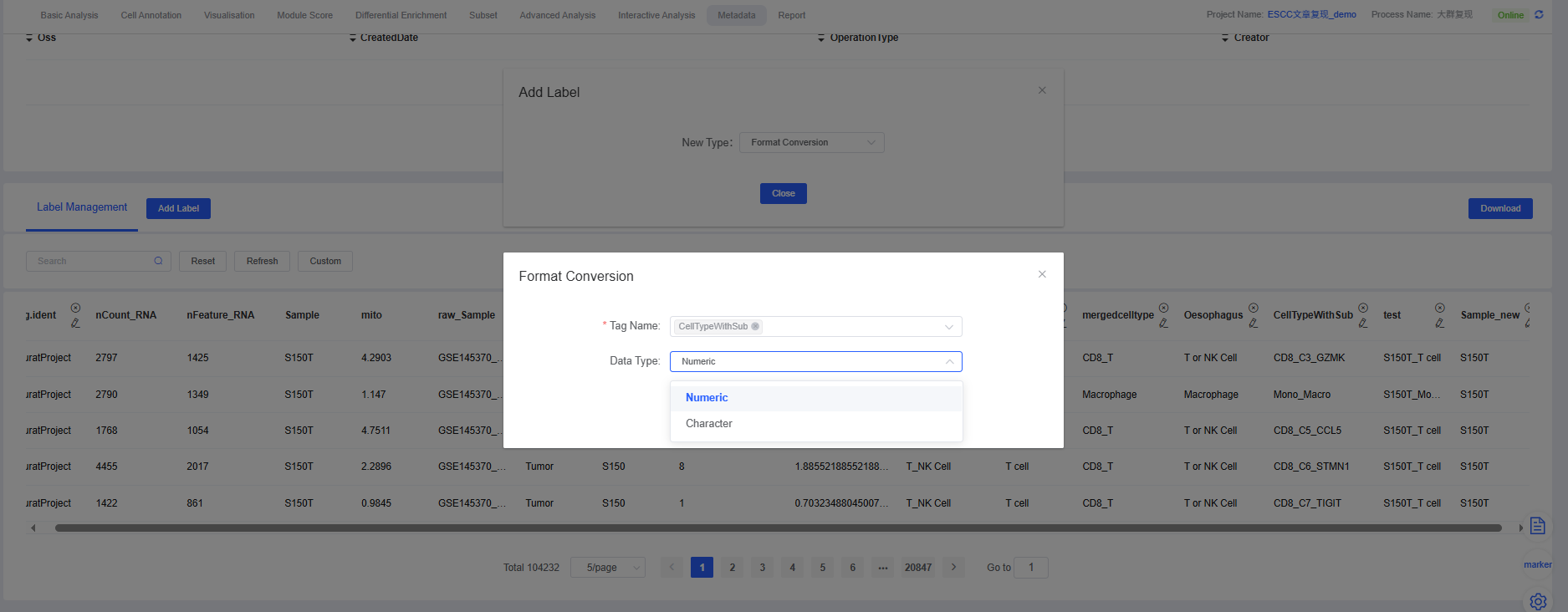
7. Format Conversion
The “Format Conversion” feature allows you to change the data type of a specific column in your metadata. The correct data type is crucial for subsequent analysis and visualization.
Common Conversion Scenarios:
- Numeric to character: Convert numeric variables (e.g., cluster IDs) to character variables. This is useful when these variables need to be treated as categorical conditions for grouping or plotting.
- Character to numeric: In scenarios where mathematical calculations are required, you may need to convert labels to numeric values.
Why is it important?
- Analysis compatibility: Certain statistical analyses or visualization tools require input data of specific types (e.g., grouping comparisons usually require character variables).
- Data interpretation: Correct formats help the platform and users correctly interpret data, avoiding the misinterpretation of categorical information as continuous values.
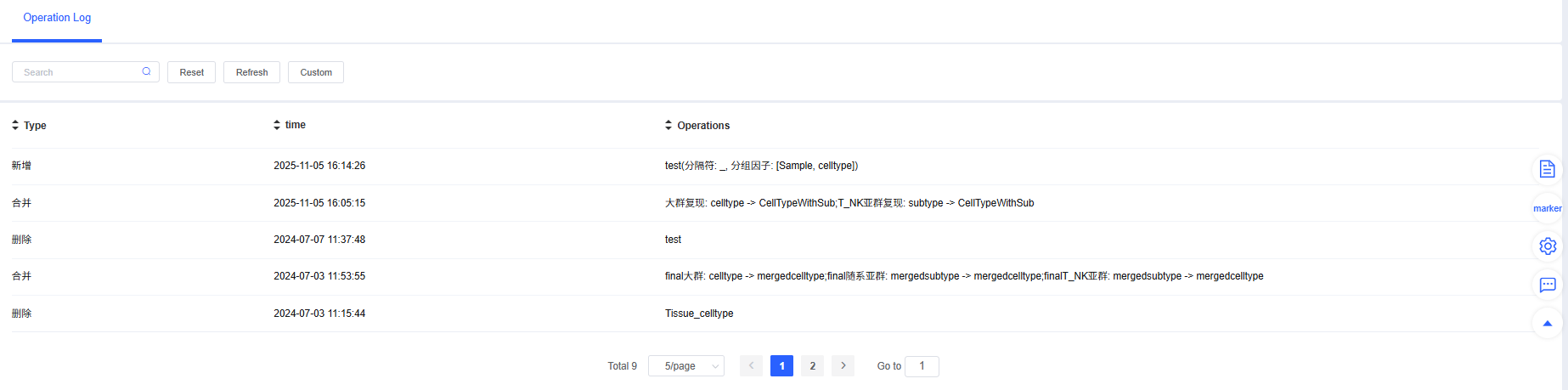
Operation Log
The “Operation Log” section is a faithful record of your project’s analysis history. It details every modification, addition, deletion, and other operations performed on meta.data, including operation times and specific content.
Core Value:
- Traceable analysis process: When you or your collaborators have questions about analysis results, you can quickly trace back through the log to understand how each label was generated or modified.
- Data security: All operations are recorded, effectively preventing accidental or malicious modifications that could lead to data chaos, ensuring the rigor of the analysis.